DataScience Workbook
/
08: Data Visualization
/ 2.
Introduction to scientific graphing
/ 2.3. RStudio – data processing & plotting with R
Introduction
Predicting the age of abalone from physical measurements.
Dataset characteristics: multivariate
Attribute type: categorical, integer, real
Subject area: life
Instances: 4177
Associated Tasks: classification
Attributes: 8
abalone <- readr::read_csv( "abalone.data", col_names = F )
names( abalone ) <- c( "Sex", "Length", "Diameter", "Height", "Whole_weight", "Shucked_weight", "Viscera_weight", "Shell_weight", "Rings" )
abalone
## # A tibble: 4,177 × 9
## Sex Length Diameter Height Whole_weight Shucked_weight Viscera_weight Shell_weight Rings
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 M 0.455 0.365 0.095 0.514 0.224 0.101 0.15 15
## 2 M 0.35 0.265 0.09 0.226 0.0995 0.0485 0.07 7
## 3 F 0.53 0.42 0.135 0.677 0.256 0.142 0.21 9
## 4 M 0.44 0.365 0.125 0.516 0.216 0.114 0.155 10
## 5 I 0.33 0.255 0.08 0.205 0.0895 0.0395 0.055 7
## 6 I 0.425 0.3 0.095 0.352 0.141 0.0775 0.12 8
## 7 F 0.53 0.415 0.15 0.778 0.237 0.142 0.33 20
## 8 F 0.545 0.425 0.125 0.768 0.294 0.150 0.26 16
## 9 M 0.475 0.37 0.125 0.509 0.216 0.112 0.165 9
## 10 F 0.55 0.44 0.15 0.894 0.314 0.151 0.32 19
## # ℹ 4,167 more rows
glimpse( abalone )
## Rows: 4,177
## Columns: 9
## $ Sex <chr> "M", "M", "F", "M", "I", "I", "F", "F", "M", "F", "F", "M", "M", "F", "F", "M", "I", "F", "M", "M", "M", "I"…
## $ Length <dbl> 0.455, 0.350, 0.530, 0.440, 0.330, 0.425, 0.530, 0.545, 0.475, 0.550, 0.525, 0.430, 0.490, 0.535, 0.470, 0.5…
## $ Diameter <dbl> 0.365, 0.265, 0.420, 0.365, 0.255, 0.300, 0.415, 0.425, 0.370, 0.440, 0.380, 0.350, 0.380, 0.405, 0.355, 0.4…
## $ Height <dbl> 0.095, 0.090, 0.135, 0.125, 0.080, 0.095, 0.150, 0.125, 0.125, 0.150, 0.140, 0.110, 0.135, 0.145, 0.100, 0.1…
## $ Whole_weight <dbl> 0.5140, 0.2255, 0.6770, 0.5160, 0.2050, 0.3515, 0.7775, 0.7680, 0.5095, 0.8945, 0.6065, 0.4060, 0.5415, 0.68…
## $ Shucked_weight <dbl> 0.2245, 0.0995, 0.2565, 0.2155, 0.0895, 0.1410, 0.2370, 0.2940, 0.2165, 0.3145, 0.1940, 0.1675, 0.2175, 0.27…
## $ Viscera_weight <dbl> 0.1010, 0.0485, 0.1415, 0.1140, 0.0395, 0.0775, 0.1415, 0.1495, 0.1125, 0.1510, 0.1475, 0.0810, 0.0950, 0.17…
## $ Shell_weight <dbl> 0.150, 0.070, 0.210, 0.155, 0.055, 0.120, 0.330, 0.260, 0.165, 0.320, 0.210, 0.135, 0.190, 0.205, 0.185, 0.2…
## $ Rings <dbl> 15, 7, 9, 10, 7, 8, 20, 16, 9, 19, 14, 10, 11, 10, 10, 12, 7, 10, 7, 9, 11, 10, 12, 9, 10, 11, 11, 12, 15, 1…
abalone %>%
select( where(is.numeric) ) %>%
psych::describe() %>%
kableExtra::kbl() %>%
kableExtra::kable_styling( bootstrap_options = c("striped", "hover") )
| vars | n | mean | sd | median | trimmed | mad | min | max | range | skew | kurtosis | se | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length | 1 | 4177 | 0.5239921 | 0.1200929 | 0.5450 | 0.5324783 | 0.1186080 | 0.0750 | 0.8150 | 0.7400 | -0.6394138 | 0.0616411 | 0.0018582 |
| Diameter | 2 | 4177 | 0.4078813 | 0.0992399 | 0.4250 | 0.4146994 | 0.0963690 | 0.0550 | 0.6500 | 0.5950 | -0.6087607 | -0.0482711 | 0.0015355 |
| Height | 3 | 4177 | 0.1395164 | 0.0418271 | 0.1400 | 0.1402498 | 0.0370650 | 0.0000 | 1.1300 | 1.1300 | 3.1265706 | 75.8953091 | 0.0006472 |
| Whole_weight | 4 | 4177 | 0.8287422 | 0.4903890 | 0.7995 | 0.7995646 | 0.5285469 | 0.0020 | 2.8255 | 2.8235 | 0.5305773 | -0.0264756 | 0.0075877 |
| Shucked_weight | 5 | 4177 | 0.3593675 | 0.2219629 | 0.3360 | 0.3439231 | 0.2349921 | 0.0010 | 1.4880 | 1.4870 | 0.7185815 | 0.5912553 | 0.0034344 |
| Viscera_weight | 6 | 4177 | 0.1805936 | 0.1096143 | 0.1710 | 0.1733193 | 0.1178667 | 0.0005 | 0.7600 | 0.7595 | 0.5914271 | 0.0809994 | 0.0016960 |
| Shell_weight | 7 | 4177 | 0.2388309 | 0.1392027 | 0.2340 | 0.2305173 | 0.1475187 | 0.0015 | 1.0050 | 1.0035 | 0.6204809 | 0.5281636 | 0.0021538 |
| Rings | 8 | 4177 | 9.9336845 | 3.2241690 | 9.0000 | 9.6410410 | 2.9652000 | 1.0000 | 29.0000 | 28.0000 | 1.1133019 | 2.3239123 | 0.0498868 |
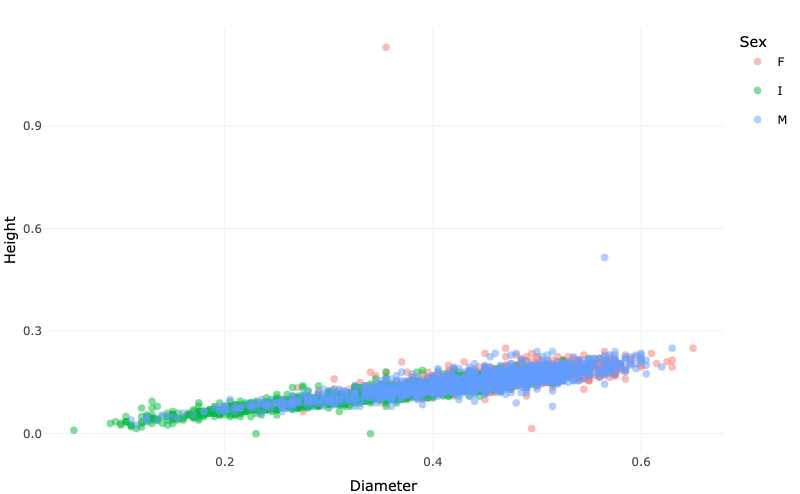
p <- ggplot(abalone, aes(x=Diameter, y=Height, color=Sex)) + geom_point() + theme_minimal()
p <- ggplotly(p)
p
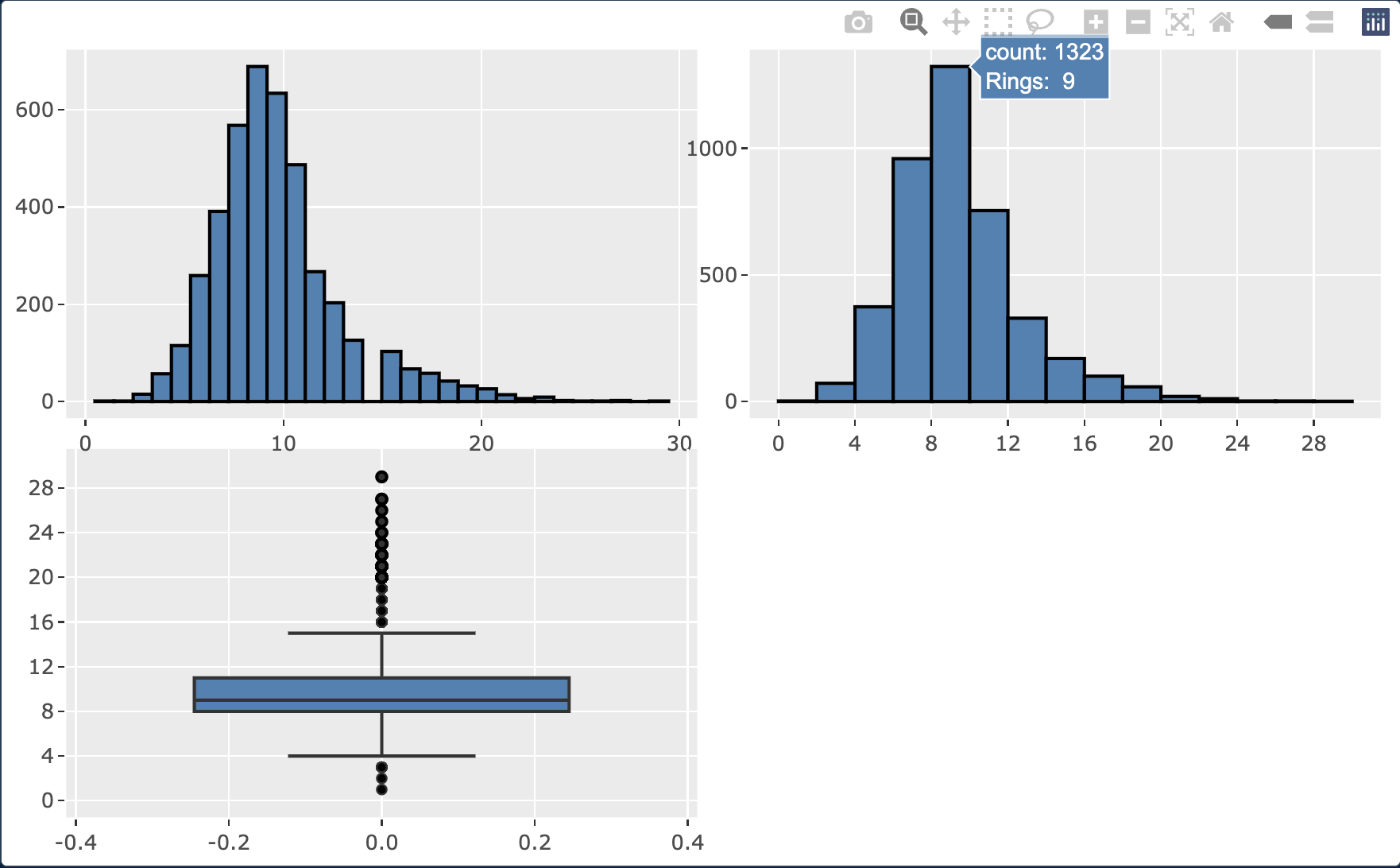
# Create a histogram with the default bin range
p1 <- abalone %>%
ggplot(aes(x=Rings)) +
geom_histogram(color="black", fill="steelblue")
# Convert to plotly
p1 <- ggplotly(p1)
# Create a histogram with a custom bin range
p2 <- abalone %>%
ggplot(aes(x=Rings)) +
geom_histogram(binwidth = 2, boundary = 0, color="black", fill="steelblue") +
scale_x_continuous(breaks = seq(0, 30, 4), limits = c(0, 30))
# Convert to plotly
p2 <- ggplotly(p2)
# Create a boxplot
p3 <- abalone %>%
ggplot(aes(y=Rings)) +
geom_boxplot(fill="steelblue") +
scale_y_continuous(breaks = seq(0, 30, 4), limits = c(0, 30))
# Convert to plotly
p3 <- ggplotly(p3)
# Print the plots using the `subplot` function from plotly
p4 <- subplot( p1, p2, p3, nrows = 2 )
p4
# Compute the correlation matrix
corr_matrix <- cor( abalone %>% select(where(is.numeric)), method = "spearman" )
# Convert the correlation matrix into a tidy format
corr_df <- as.data.frame( corr_matrix ) %>%
rownames_to_column( "Var1" ) %>%
reshape2::melt( id.vars = "Var1", variable.name = "Var2", value.name = "Correlation" )
# Create a heatmap with ggplot2
heatmap <- ggplot( corr_df, aes(x = Var1, y = Var2, fill = Correlation) ) +
geom_tile() +
geom_text( aes(label = round(Correlation, 2)), size = 4 ) +
scale_fill_gradient2( low = "blue", high = "red", mid = "white", midpoint = 0 ) +
theme_minimal() +
theme( axis.text.x = element_text(angle = 45, hjust = 1) )
# Convert the heatmap to a plotly interactive graphic
heatmap <- ggplotly(heatmap)
heatmap
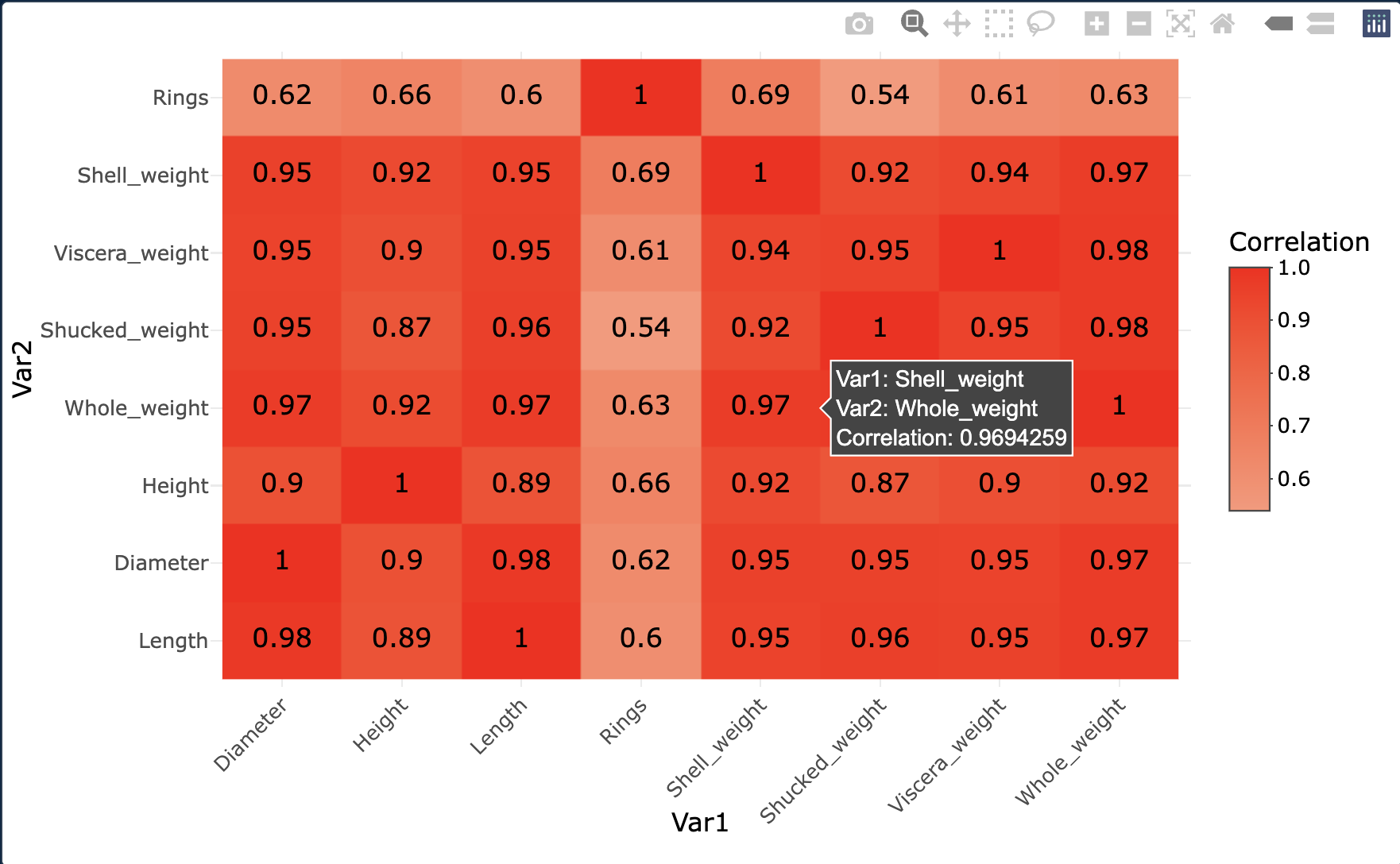
The attributes that are most correlated to Rings are Shell_weight
and Height.