Introduction
Sometimes you may need to download a specific folder from a repository hosted on GitHub, but does not want to download the entire repository due to its large size. For example, you may only need a particular folder that contains documentation or script related to a specific project.
Downloading a single folder can be particularly useful if the repository is large and contains a lot of files that are not relevant to the task at hand, as cloning the entire repository can be time-consuming and take up a lot of storage space on the local machine. In this case, the users can download only the specific folder that they need, rather than the entire repository, using:
-
SVN version control system, to download from the command line
This can be done on any local or remote machine, including HPC systems. -
online code editors, such as built-in GitHub VS Code ⤴ or CodeSandbox ⤴, to download in the browser
This can be done on a local machine and HPC cluser that has the Open OnDemand interface.
These approches can save time and resources and allow the developer to work more efficiently.
Download using CLI
(command line on a local or remote machine)
Get started with SVN
Subversion (SVN) is a popular open-source centralized version control system that allows developers to manage and track changes to their codebase. SVN is widely used in software development, as it allows teams to collaborate and maintain a single, up-to-date version of their codebase.
SVN can be considered as an alternative for Git (distributed version control tool) and may be a better choice for smaller teams or for organizations with a more centralized development structure.
…about version control systems such as SVN or Git, follow the tutorials provided in section 09. Project Management / Storage & Version Control.
To get started with SVN, check if it pre-installed on your computing machine, either local one or remote HPC system. In the terminal window type the following command:
svn --version
This command checks if SVN is installed and displays the version number of SVN.
^ if it is installed on the system
svn, version 1.14.3 (r1914484) compiled Dec 9 2023, 13:27:10 on arm-apple-darwin23.2.0 Copyright (C) 2023 The Apache Software Foundation. This software consists of contributions made by many people; see the NOTICE file for more information. Subversion is open source software, see http://subversion.apache.org/
If the command is not recognized, it means that SVN is not installed on the system.
svn: command not found
If SVN is not installed on the HPC system, you may need to contact your system administrator to request that it be installed.
If SVN is not installed on your local machine, you will first need to download and install it on your system. SVN is available for Windows, macOS, and Linux, and can be downloaded from the Apache Subversion website: https://subversion.apache.org/download/ ⤴.
Ubuntu/Debian:
Open a terminal and run the following command to update the package list.
Then install SVN by running the following command:
sudo apt-get update
sudo apt-get install subversion
Windows:
- Download the SVN client for Windows from the official Apache website, https://www.apache.org/dyn/closer.lua/subversion/ ⤴.
- Choose the appropriate version of SVN for your Windows system (32-bit or 64-bit) and click on the download link to start the download.
- Once the download is complete, double-click on the downloaded file to start the installation.
- Follow the on-screen instructions to complete the installation.
macOS:
Install homebrew ⤴ if you don’t already have it.
Then install SVN using Homebrew by running the following command in a terminal:
brew install svn
Once you have installed SVN, you can start using it!
GitHub Folder
One common use case is to download a single folder from a GitHub repository or other remote repository using the svn export command.
- First, navigate to the location in your file system where you want to download.
You can do this by using thecdcommand in your terminal to navigate to the parent folder, and then using thelscommand to list the contents of the folder.cd path/to/destination/location -
Go to the online repository and find a folder you want to download. Copy the URL from the address bar in the web browser.
Let’s use the bin_data folder in the https://github.com/ISUgenomics/data_wrangling ⤴ GitHub repo as an example: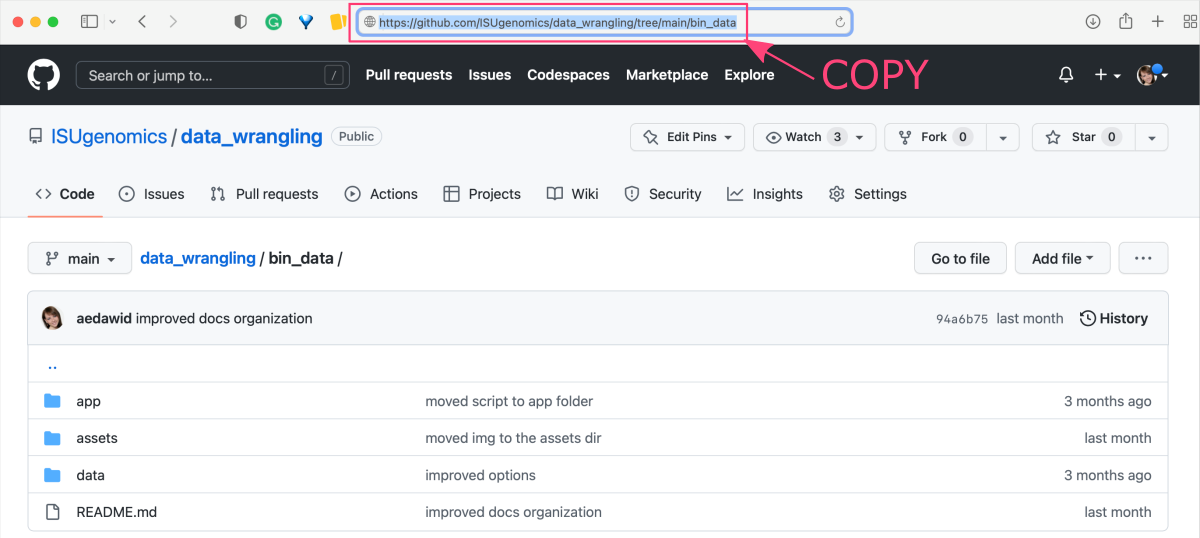
- Now, go back to the terminal window to download only the selected folder.
First, typesvn exportand paste the copied URL:svn export https://github.com/ISUgenomics/data_wrangling/tree/main/bin_dataNOTE: Before you execute the command, replace
tree/mainwithtrunkkeyword.
Now, you can run the command:svn export https://github.com/ISUgenomics/data_wrangling/trunk/bin_data
3’. To make it easier for future use, create an empty script file, e.g., get_folder.sh and copy-paste the code snippet:
#!/bin/bash
echo "----------------"
echo "USAGE:"
echo " . ./get_GitHub_folder.sh <URL to the GitHub folder>"
echo " . ./get_GitHub_folder.sh https://github.com/ISUgenomics/common_scripts/tree/master/get_GitHub_file"
echo "----------------"
echo ""
URL=`echo $1`
folder=`echo $URL | sed 's|tree/master|trunk|g'`
svn export $folder
Make the script executable:
chmod u+rwx get_folder.sh
From now on, you do not need to replace keywords in the copy-pasted URL because it will be done automatically using the script.
So, to download the folder from the example above, simply type in the command line:
get_folder.sh https://github.com/ISUgenomics/data_wrangling/tree/main/bin_data
Ideally, you should place the script in a directory with your all universal scripts, such as ~/SCRIPTS. This way the path will be easy to remember. You can also add it to the $PATH environment variable to make it easier to execute the script.
export PATH=$PATH:~/SCRIPTS
For this change to be applied to every shell you open, add it to the file that the shell will source when it is invoked.
source ~/.bashrc
Then you can execute the script by its name from any location in the file syetem:
get_folder.sh https://github.com/ISUgenomics/data_wrangling/tree/main/bin_data
GitHub File
If you need to download a single file from the GitHub repository you can use svn export or wgetcommand in the terminal. This solution can be useful if you want to have the file downloaded directly to the remote machine such as HPC cluster.
use wget command
The simplest solution to download a single file from the GitHub repository is the wget command.
You can use the wget command to download any type of file from online repository.
-
Open selected GitHub repository in any web browser and navigate to the file that you want to download.
-
Click the file name to open the preview of the GitHub rendering. Then, click the
Rawbutton (top-right corner) to open the source code of the file.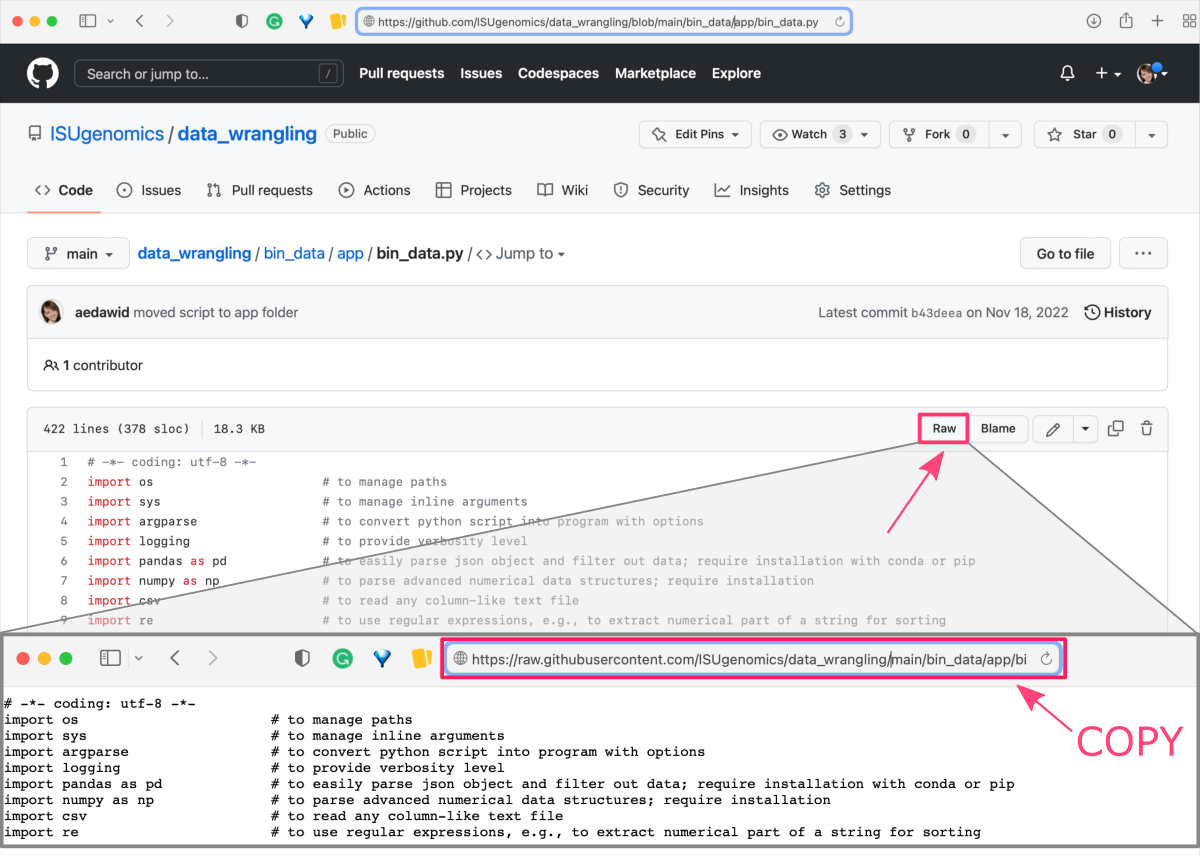
-
Copy the URL address of the raw file.
-
Open the terminal window and navigate to the desired location in the file system (on a local or remote machine).
-
Use the
wgetcommand followed by the copied URL:wget https://raw.githubusercontent.com/ISUgenomics/data_wrangling/main/bin_data/app/bin_data.py
use svn export command
To get started with the SVN tool (e.g., to check if you have it installed) see section Get started with SVN in this tutorial. To learn more about version control systems, including SVN and Git, explore tutorials in section 09. Project Management / Storage & Version Control of this workbook.
Once you have installed SVN, you can use it to download a single file from the GitHub!
- First, navigate to the location in your file system where you want to download.
You can do this by using thecdcommand in your terminal to navigate to the parent folder, and then using thelscommand to list the contents of the folder.cd path/to/destination/location -
Go to the online repository, find a file you want to download and click on it to open its preview. Copy the URL from the address bar in the web browser.
As an example, let’s use thebin_data.pyfile located in the bin_data/app ⤴ folder of the data_wrangling GitHub repo: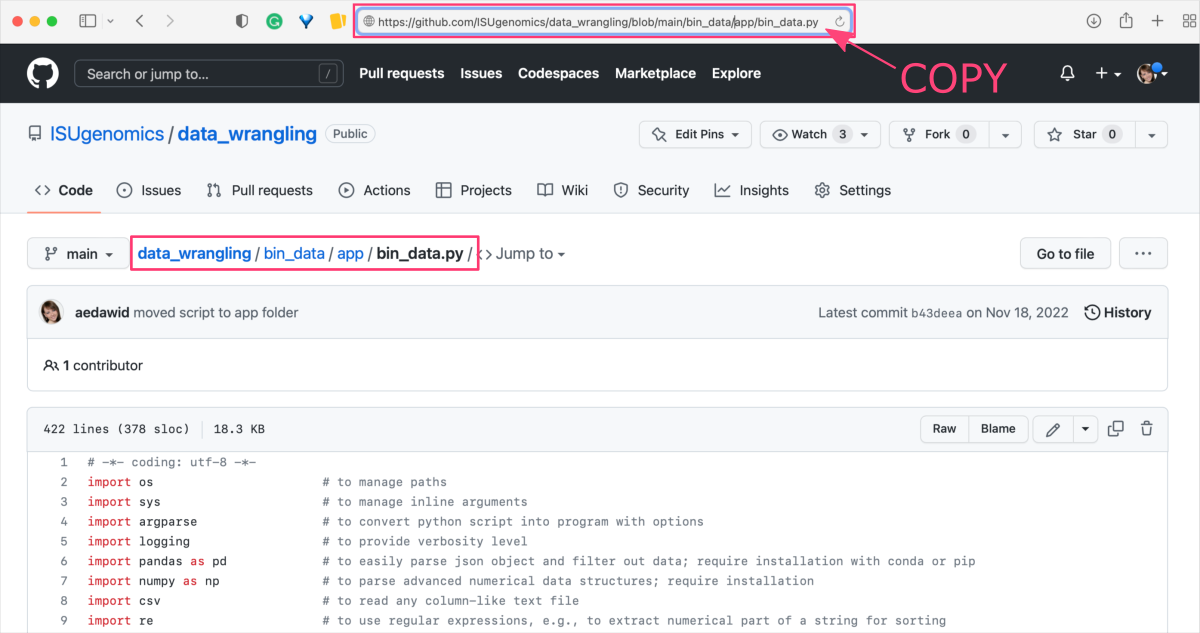
- Now, go back to the terminal window to download only the selected file. First, type
svn exportand paste the copied URL.svn export https://github.com/ISUgenomics/data_wrangling/blob/main/bin_data/app/bin_data.pyNOTE: Before you execute the command, replace
blob/masterorblob/mainwithtrunkkeyword.svn export https://github.com/ISUgenomics/data_wrangling/trunk/bin_data/app/bin_data.py
3’. To make it easier for future use, create an empty script file, e.g., get_file.sh and copy-paste the code snippet:
#!/bin/bash
echo "----------------"
echo "USAGE:"
echo " . ./get_GitHub_file.sh <URL to the GitHub file>"
echo "e.g., . ./get_GitHub_file.sh https://github.com/ISUgenomics/common_scripts/blob/master/get_GitHub_file/get_GitHub_file.sh"
echo "----------------"
echo ""
URL=`echo $1`
file=`echo $URL | sed 's|blob/master|trunk|g' | sed 's|blob/main|trunk|g'`
svn export $file
get_GitHub_file.sh (END)
Make the script executable:
chmod u+rwx get_file.sh
From now on, you do not need to replace keywords in the copy-pasted URL because it will be done automatically using the script.
So, to download the file from the example above, simply type in the command line:
get_file.sh https://github.com/ISUgenomics/data_wrangling/blob/main/bin_data/app/bin_data.py
Download from a browser
(manual download from the web-based GUI)
These approach require browser access directly on the machine where the download will take place.
If you want to download a single folder from a GitHub repository to a remote HPC system that doesn’t have a browser-based GUI, such as Open On Demand, then you need to
1) download the folder to a local machine and further
2) transfer it to the cluster
using, for example, an ssh connection and the scp or rsync commands.
GitHub Folder
Edge | Opera | Chrome | Chromium
If you have one of the browsers: Edge, Opera, Chrome, or Chromium…
-
use it to open the GitHub repository of your choice, and
-
navigate to the folder in the repo that you want to download.
-
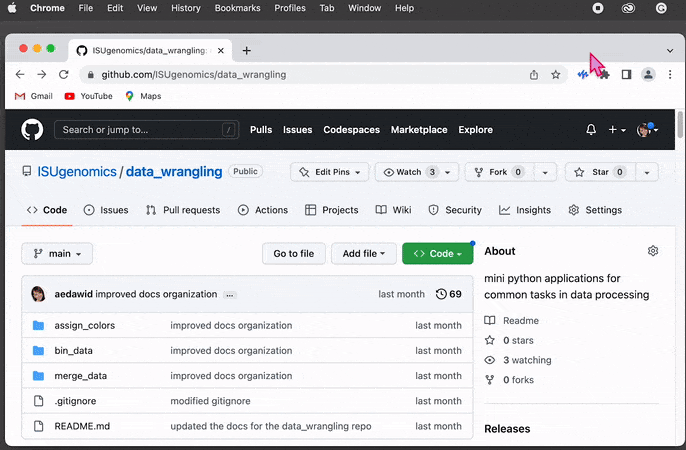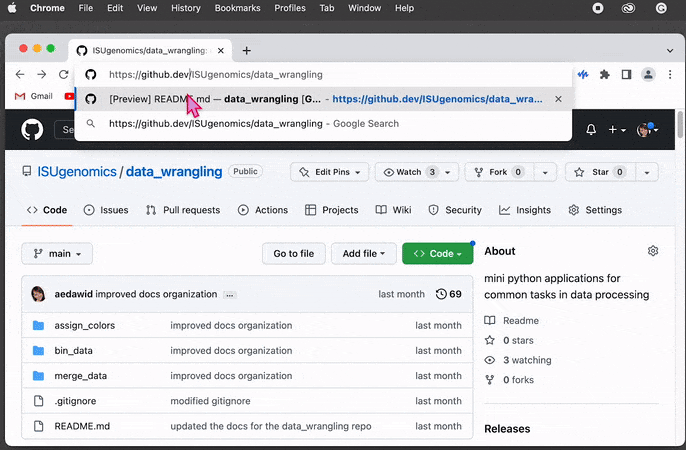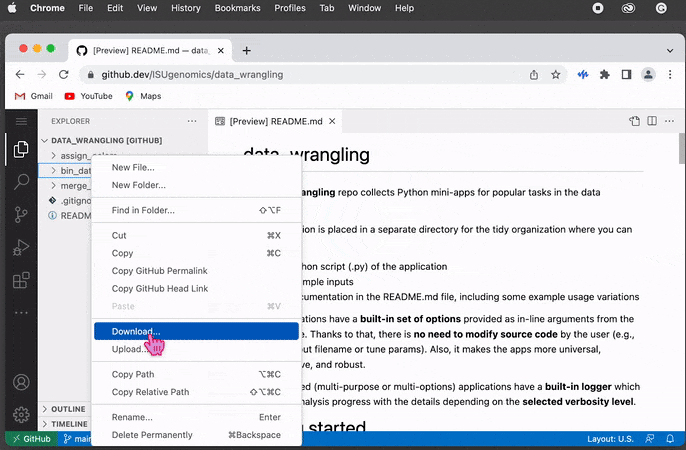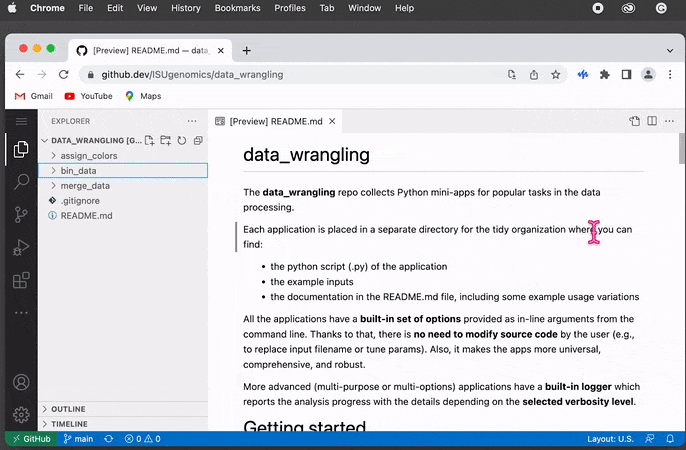
Press
.(dot) on your keyboard or replace manually.comwith.devin URL
This will open the repo in the GitHub’s internal editor directly in the browser window -
On the left-hand side, you can see the
Explorermenu. Find the folder andright-clickon it to display the dialog box. Then click on theDownloadoption and select the directory on your file system as a location to save the content.
Any browser: Safari, Firefox
So far, the most robust and easiest way to download a single folder from a GitHub repository is to do it through an online editor such as codesandbox, which works reliably regardless of browser.
-
Open selected GitHub repository in any web browser and navigate to the folder in the repo that you want to download
-
In the URL address, replace github with githubbox
This will open the repo in the codesandbox online editor directly in the browser window -
On the left-hand side, you can see the File System browser. Find the folder and
clickon the Export to ZIP option . This will download the zipped folder automatically and save it in your default location.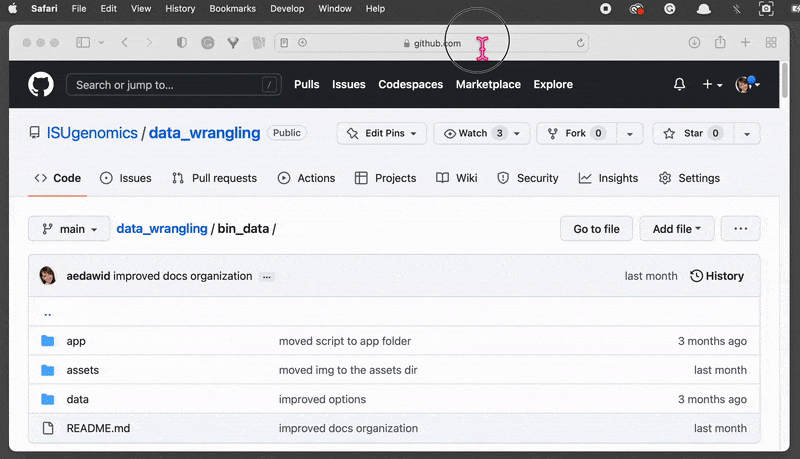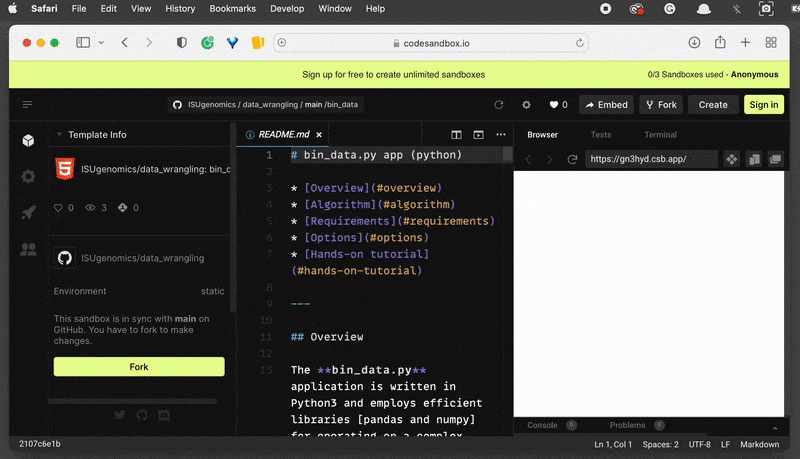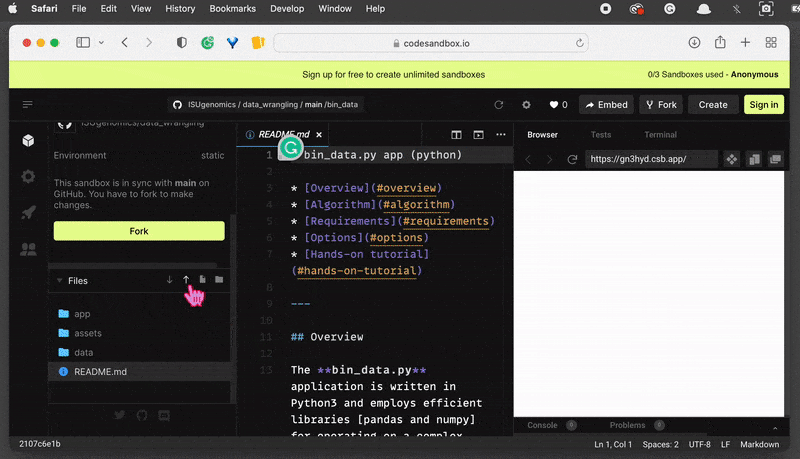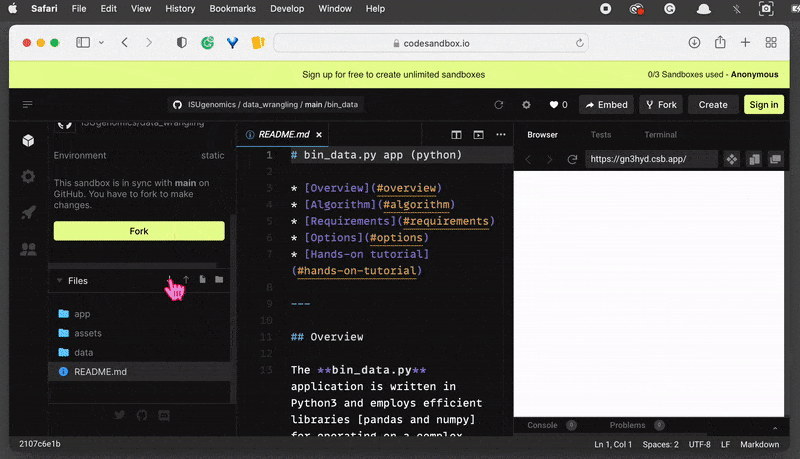
To decompress the archive, use
unziportar -xfcommand followed by the name of the downloaded file:unzip isugenomics-data-wrangling-bin-data.zip
GitHub File
If you need to download a single file from the GitHub repository to your local machine, you can do that directly from a web browser.
-
Open selected GitHub repository in any web browser and navigate to the file that you want to download.
-
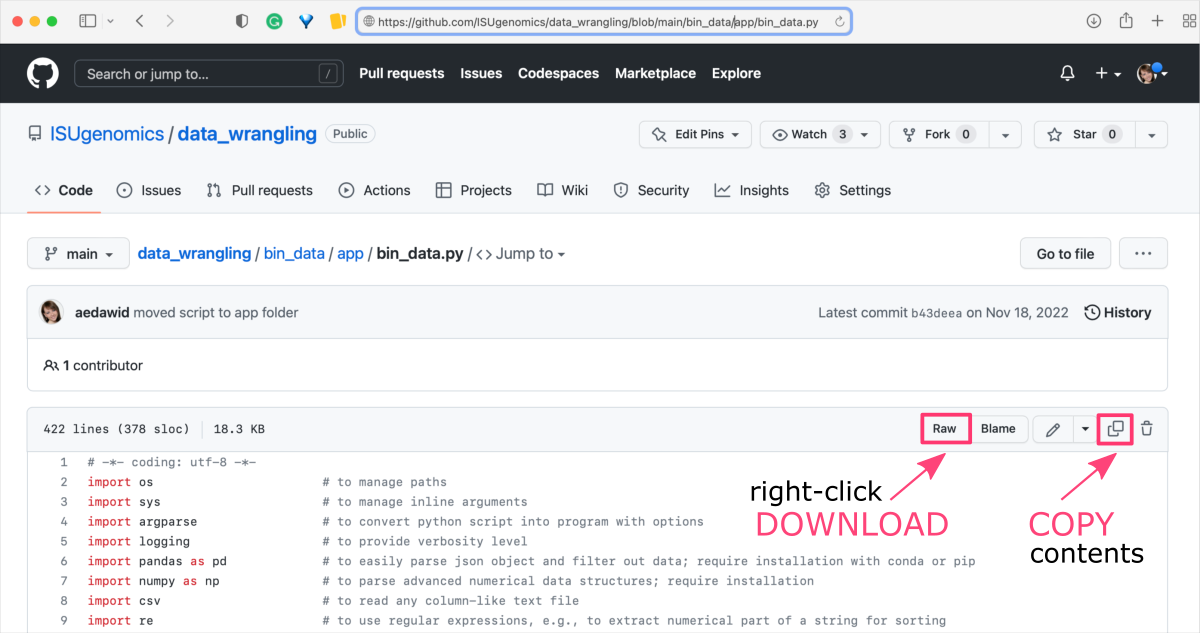
Click the file name to open the preview of the GitHub rendering. Then, in the upper right corner you should see the horizontal menu with several buttons, including
Raw,Blame,Edit,Copy raw contents, andDelete this file.- Right-click on the
Raw buttonand selectDownload linked Filefrom the pop-up dialog box.
Navigate to the location where you save downloads by default to find the file. or - Click-on the
Copy raw contentsbutton to copy the contents of the file to the clipboard.
You can then paste the copied contents into any text file.
- Right-click on the
Further Reading
Remote data preview (without downloading)Viewing text files using UNIX commands
Viewing PDF and PNG files using X11 SSH connection
Viewing graphics in a terminal as the text-based ASCII art
Mounting remote folder on a local machine
Data manipulation
Data wrangling: use ready-made apps
MODULE 08: Data Visualization
